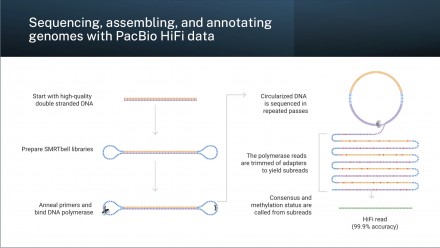
Sequencing, assembling, and annotating genomes with PacBio HiFi data
Workshop participants will gain knowledge and hands-on experience in analysing PacBio HiFi sequencing data, running whole genome assemblies, evaluating the quality of their genome assemblies, and will be introduced to whole genome annotation.
Target audience: students, postdocs, and other researchers who are interested in using whole genome sequencing in their research.
Participants will need to bring their own laptops with an ssh application installed (further details will be provided prior to the workshop).
Day 1: Tues 18 FebMorning:
Afternoon:
Day 2: Wed 19 FebMorning:
Afternoon:
|
About Paul
Paul Frandsen is interested in evolutionary genomics and phylogenetics. His most recent work has focused on the genomic basis of silk production in caddisflies and lepidopterans and the use of genetic techniques in biodiversity monitoring. He also has considerable expertise in genome sequencing, assembly, and annotation.
Paul is visiting ANU in February 2025. His visit is funded by the Centre for Biodiversity Analysis, hosted Rob Lanfear (ANU). Please get in touch with Rob if you would like to meet with Paul during his time in Canberra.
Paul will also be giving a seminar at ANU on Thursday 20 February: Threading links between the silk genotype and phenotype in caddisflies, nature’s underwater architects.