TreeScaper: Visualizing and Extracting Phylogenetic Signal from Sets of Trees
Modern phylogenomic analyses often result in large collections of phylogenetic trees representing uncertainty in individual gene trees, variation across genes,
Speakers
Event series
Content navigation
RegisterDescription
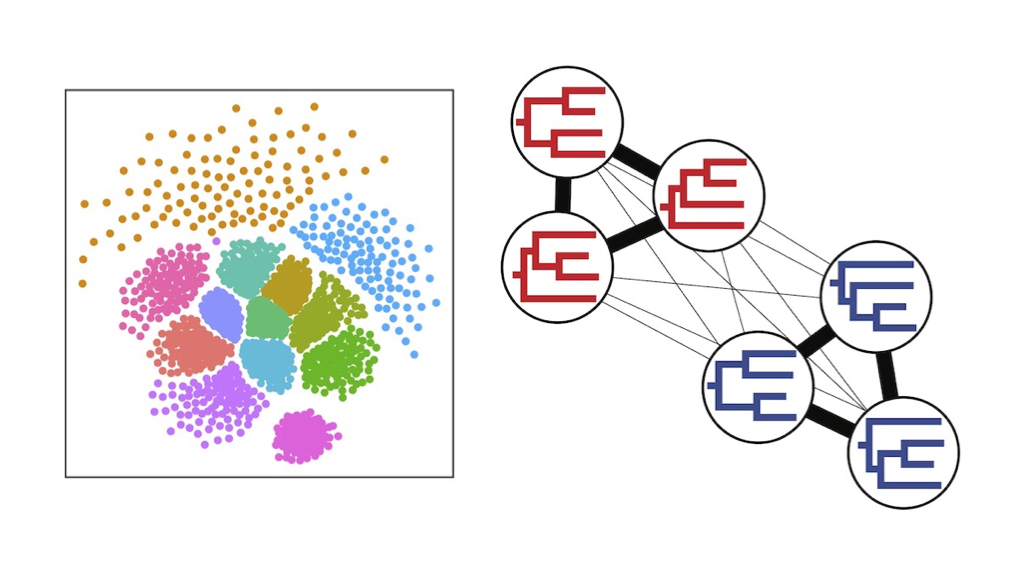
Modern phylogenomic analyses often result in large collections of phylogenetic trees representing uncertainty in individual gene trees, variation across genes, or both. Extracting phylogenetic signal from these tree sets can be challenging, as they are difficult to visualize, explore, and quantify.
This workshop will cover the use of TreeScaper to address some of these challenges. TreeScaper consists of two fundamental sets of tools:
- Visualization methods to explore tree space in 2- or 3- dimensions; and
- Community detection methods that utilize networks to look for distinct phylogenetic signals.
This workshop will cover background on the methods available in TreeScaper, as well as installation and use of the software.
The workshop is free but please register if you would like to attend as places are limited.
Participants should bring their own laptop running either Mac OS or Linux, and any sets of phylogenetic trees that they want to try and analyze.
Please download and install TreeScaper prior to the workshop. Below are a few notes about installing TreeScaper, particularly for those downloading the executable version of the program on a Mac:
- After downloading and unzipping the file, you will need to double click on TreeScaper.dmg. This will mount a disk image containing TreeScaper. Drag the program back to the folder you downloaded.
- On a Mac, when you try and run TreeScaper for the first time by double clicking, it will produce an error and warn you that the program is from an “unidentified developer”. To get around this, hold down option and then click once. From the menu that pops up, select “Open”. Confirm that you want to continue opening the program. After you do this once, you should be able to run the program just by double clicking on it.
- TreeScaper relies on external files to store settings. Therefore, the program needs to be in the same folder as these files to run properly. If you want to put the program in your Applications directory, please move the entire folder that you downloaded.
If you have problems with installation, please let Jeremy know as soon as possible.
Further reading: Wen Huang, Guifang Zhou, Melissa Marchand, Jeremy R. Ash, David Morris, Paul Van Dooren, Jeremy M. Brown, Kyle A. Gallivan, Jim C. Wilgenbusch 2016. TreeScaper: Visualizing and Extracting Phylogenetic Signal from Sets of Trees. Molecular Biology and Evolution 33(12): 3314-3316. https://doi.org/10.1093/molbev/msw196
Location
DNA Meeting Room (105), Level 1, RN Robertson Building, ANU




