SNPs in population- and phylo-genomics
Have you got large SNP datasets and now are wondering how best to filter the data, to infer population diversity or structure, adaptation, or to delimit species
Speakers
Event series
Content navigation
RegisterDescription
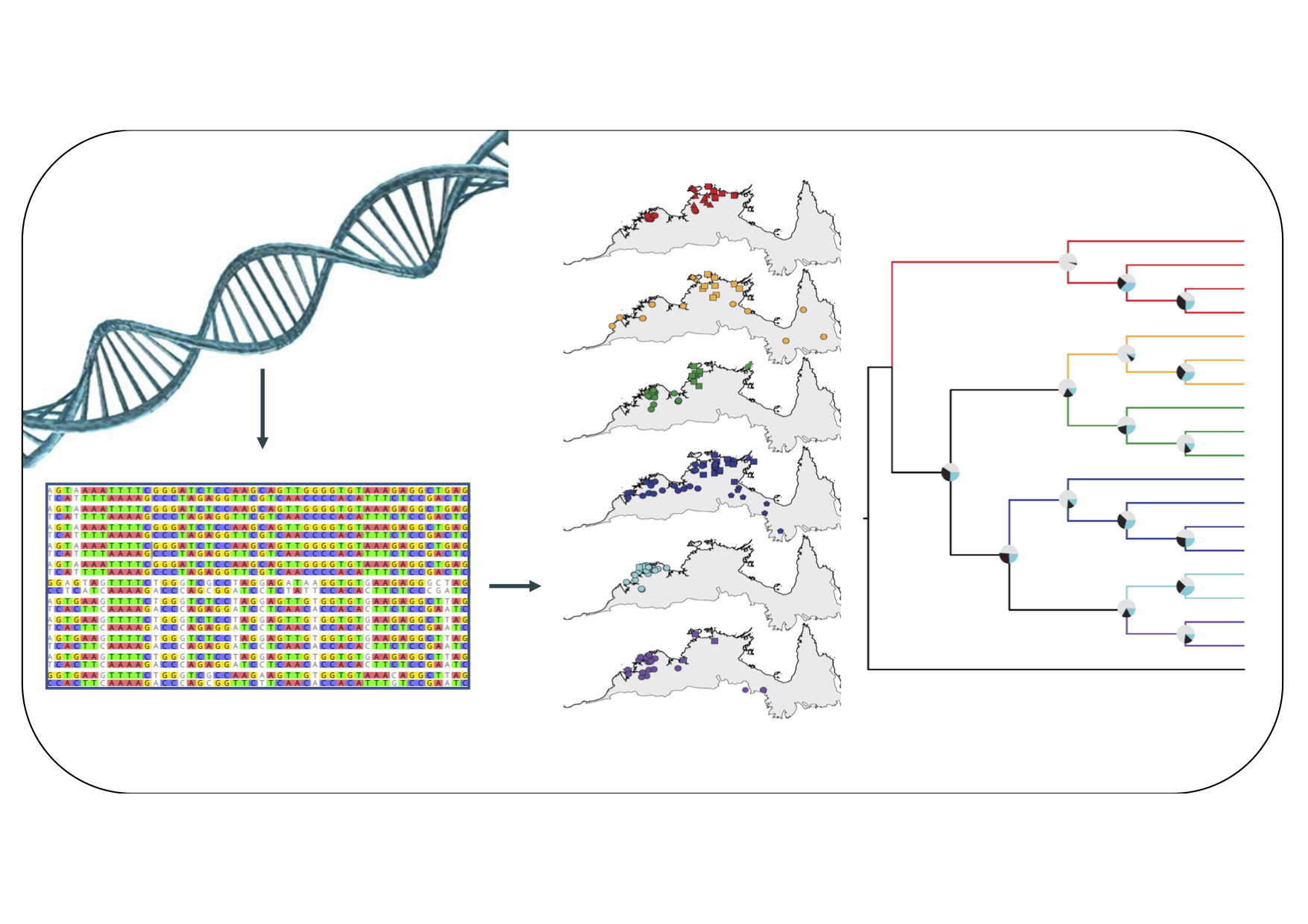
Have you got large SNP datasets and now are wondering how best to filter the data, to infer population diversity or structure, adaptation, or to delimit species and estimate their relationships? Do you have a bioinformatics or analytical pipeline that you would like to see used by empiricists?
With increasing access to large-scale SNP data (e.g. from ddRAD, GbS or DArT screens), many of us now need to understand the nuances of cleaning and filtering data, and then analyzing the data.
This 3-day workshop will include lectures and hands-on training from Australian and international experts spanning bioinformatics, population genomics and phylogenetics with SNP data.
Emphasis will be on applications for non-model species, but there will also be examples of how to exploit genome assemblies from related taxa should these be available. There will also be opportunity for both empiricists and developers of methods to present posters in order to build a community that can continue to interact to get the most out of such datasets.
Registration: Closes 22 November, 2018. Register online here.
Please note - registration has now closed and the workshop is at capacity.
Participants will need to bring their own laptops. More detailed instructions regarding software, data, etc. will be emailed to prior to the workshop. A pre-workshop survey for registered participants can be found here.
ECR grants: The CBA is also offering several Early Career Researcher grants of $150 AUD each to cover registration costs. Please note in your application if you would like to be considered for one of these.
Cost: $150 for 3 days (includes all morning and afternoon teas and lunches, and drinks on Tuesday evening). A social function (Mexican food and drinks) will also be held on the Wednesday evening ($45 per head).
Venue maps & information:
- University of Canberra map
- Venue map
- Casual parking on campus*
- Short-term accomodation on campus
* Free workshop parking has been arranged at parking areas P7 & P17 (see map). Press the intercom button at the gate, state your name and that you are a “CBA workshop participant”. Then see Bernd Gruber at the workshop who will give you a ticket to leave the car park.
ProgramDownload Program |
|
TUESDAY 4 DECEMBER 2018 |
|
Introduction |
|
|
9:00 |
LECTURE Welcome & overview on applications of SNP data - Craig Moritz (slides) |
| 9:45 | LECTURE DArT pipeline - Andrzej Kilian (slides) |
| 10:15 | LECTURE Population genomics introduction - Sam Banks (slides) |
| 10:45 | Morning tea break |
|
11:00 |
Filtering SNPs with dartRComputer setup dartR (Groups A & B) dartR requirements for workshop |
| 11:15 | TUTORIAL (Groups A & B) Introduction to dartR - quality filtering, data structures - Arthur Georges/Bernd Gruber/ Renee Catullo |
| 13:00 | Lunch (20 min DArT tours in groups of 10) |
| 14:00 | TUTORIAL (Group A) dartR introduction cont. (basic analyses) - Bernd Gruber (tutorial downloads) |
| 14:00 | TUTORIAL (Group B) QA & filtering your own data - Sam Banks & Renee Catullo (tutorial downloads) |
| 15:15 | Afternoon tea break |
| 15:45 | TUTORIAL (Group A) QA & filtering your own data - Sam Banks & Renee Catullo (tutorial download) |
| 15:45 | TUTORIAL (Group B) dartR introduction cont. (basic analyses) - Bernd Gruber (tutorial downloads) |
| 17:00 | KDDArT informatics - Andrzej Kilian (slides) |
| 18:00 |
Welcome mixer / CBA end-of-year drinks. All welcome.
|
WEDNESDAY 5 DECEMBER 2018 |
|
Population genomics |
|
| 9:00 | LECTURE Population structure & landscape genetics - Bernd Gruber (slides) |
| 9:45 | LECTURE Adaptation, GWAS & GEA - Luciano Beheregaray (slides) |
| 10:30 | Morning tea break |
| 11:00 | TUTORIAL (Group A) Population structure & landscape genetics - Bernd Gruber (tutorial downloads) |
| 11:00 | TUTORIAL (Group B) Adaptation, GWAS & GEA - Chris Brauer (tutorial downloads) |
| 12:30 | Lunch (poster presentations) |
| 13:30 | TUTORIAL (Group A) Adaptation, GWAS & GEA - Chris Brauer (tutorial downloads) |
| 13:30 | TUTORIAL (Group B) Population structure & landscape genetics - Bernd Gruber (tutorial downloads) |
| 15:00 | Afternoon tea break |
| 15:30 | LECTURE Adaptation genomics - Justin Borevitz (slides) |
| 16:15 | LECTURE dartR phylogeny - Arthur Georges (slides) |
| 17:30 | Social function (Mexican dinner & drinks), Coopers Lodge Rooftop. Pre-paid ticket-holders only. |
THURSDAY 6 DECEMBER 2018 |
|
Systematics |
|
| 9:00 | LECTURE Species tree inference - theory - Remco Bouckaert (slides) |
| 9:45 | LECTURE Species tree inference - examples - Adam Leache (slides) |
| 10:30 | Morning tea break |
| 11:00 | TUTORIAL Species tree inference - SNAPP demonstration - Adam Leache/Remco Bouckaert |
| 12:30 | Lunch |
| 13:30 | LECTURE Species delimitation - theory & examples - Adam Leache/Remco Bouckaert (slides) |
| 15:00 | Afternoon tea break |
| 15:30 | TUTORIAL Species delimitation - Adam Leache/Remco Bouckaert |
| 17:30 |
Workshop ends We would appreciate you filling in our post-workshop survey. Workshop photo. |
|
|
|
Location
Building 6, Level B, Rooms 3 & 4, University of Canberra, Bruce, ACT




