Estimating biogeographic history with BioGeoBEARS and PhyBEARS
Developer Nick Matzke provides an introduction to his two powerful software packages for estimating historical biogeography.
Speakers
Event series
Content navigation
RegisterDescription
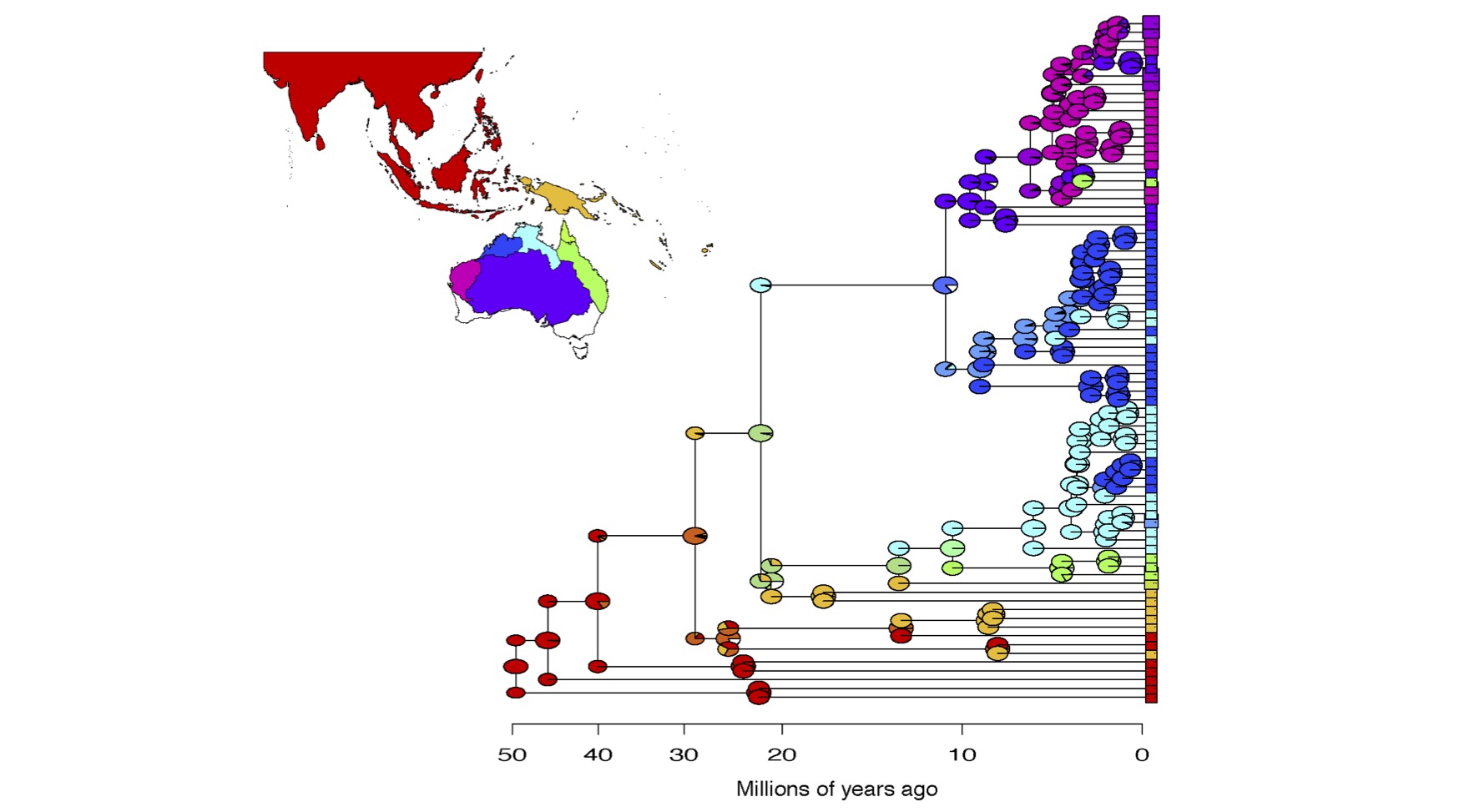
Historical biogeography methods have long been dominated by the "dispersal versus vicariance" debate, and different computer programs (e.g. DIVA, Lagrange, BayArea) have made different fixed assumptions about the importance of these processes. Researchers typically just run the different programs, and observe whether or not the inferences differ, but they have no ability to judge, statistically, which of the models best fits the data, or whether another model (for example, one including founder-event speciation) might be better than any of these.
The R package BioGeoBEARS, developed by Nicholas Matzke, allows users to build models that give different probability to vicariance, dispersal, and other processes, as well as build more complex models (dispersal probability as a function of distance, island emergence and submergence, inclusion of fossil data). All of the models are directly comparable in the common framework of statistical model choice.
However, all biogeographic reconstruction models based on discrete areas are computationally limited by the resulting number of range states, which grows exponentially with the number of areas. The standard biogeography models have also been criticized for ignoring the process of lineage extinction. In the last decade another kind of models, the State-dependent Speciation and Extinction (SSE) models, have been developed incorporating birth-death processes where the speciation and extinction rates are dependent on the state of an evolving character (such as the distribution range).
The package PhyBEARS, also developed by Matzke, makes SSE-type models accessible for large numbers of states/ranges. It is implemented in Julia, a more computationally efficient language than R. PhyBEARS can run the same models as BioGeoBEARS more efficiently, and then add lineage extinction, as well as more variants such as extinction dependent on area size.
Aims
This workshop aims to bring cutting-edge knowledge on biogeography phylogenetic comparative methods to the research community in Canberra and other researchers attending the AES conference in the following days.
Attendees of this workshop will learn how to build and test models in BioGeoBEARS and PhyBEARS, as well as necessary skills in R, Julia and statistical model selection.
Target audience
Researchers, particularly early-career ones, interested in historical biogeography and ancestral range estimation. Some previous experience in R or other programming language is preferable.
With the Australasian Evolution Society 2022 meeting at ANU in the same week (14-16 December), we welcome attendees coming from institutions across Australia and New Zealand.
Registration
Please register by 25 November. If the workshop is over-subscribed we may need to select people based on their interests and skills, and to maximise the diversity of research groups.
Successful applications will be notified by 28 November. Further workshop details will be sent then also.
Program
Monday 12 December (BioGeoBEARS, R) |
|
|
9:30-10:45 |
Introduction to BioGeoBEARS |
|
10:45-11:00 |
Morning tea (Robertson Tea Room) |
|
11:00-12:00 |
Running examples of BioGeoBEARS and model comparison |
|
12:00-13:15 |
Lunch (not provided, but plenty of options on campus) |
|
13:15-15:15 |
Bayesian Stochastic Mapping |
|
15:15-15:30 |
Afternoon tea (Robertson Tea Room) |
|
15:30-16:30 |
Advanced customised options in BioGeoBEARS |
|
|
|
Tuesday 13 December (PhyBEARS, Julia) |
|
|
9:30-10:45 |
Introduction to Julia language / setup |
|
10:45-11:00 |
Morning tea (Robertson Tea Room) |
|
11:00-12:00 |
Introduction to PhyBEARS |
|
12:00-13:15 |
Lunch (not provided, but plenty of options on campus) |
|
13:15-15:15 |
Model comparison with PhyBEARS |
|
15:15-15:30 |
Afternoon tea (Robertson Tea Room) |
|
15:30-16:30 |
Advanced topics |
Location
Slayter Seminar Room, 2nd floor, RN Robertson Building, Australian National University, Canberra




