Multi-species Coalescent Analysis
In this workshop Adam Leaché, from the University of Washington, will give an introduction and run through practical activities on coalescent theory and analysis.
Speakers
Event series
Content navigation
RegisterDescription
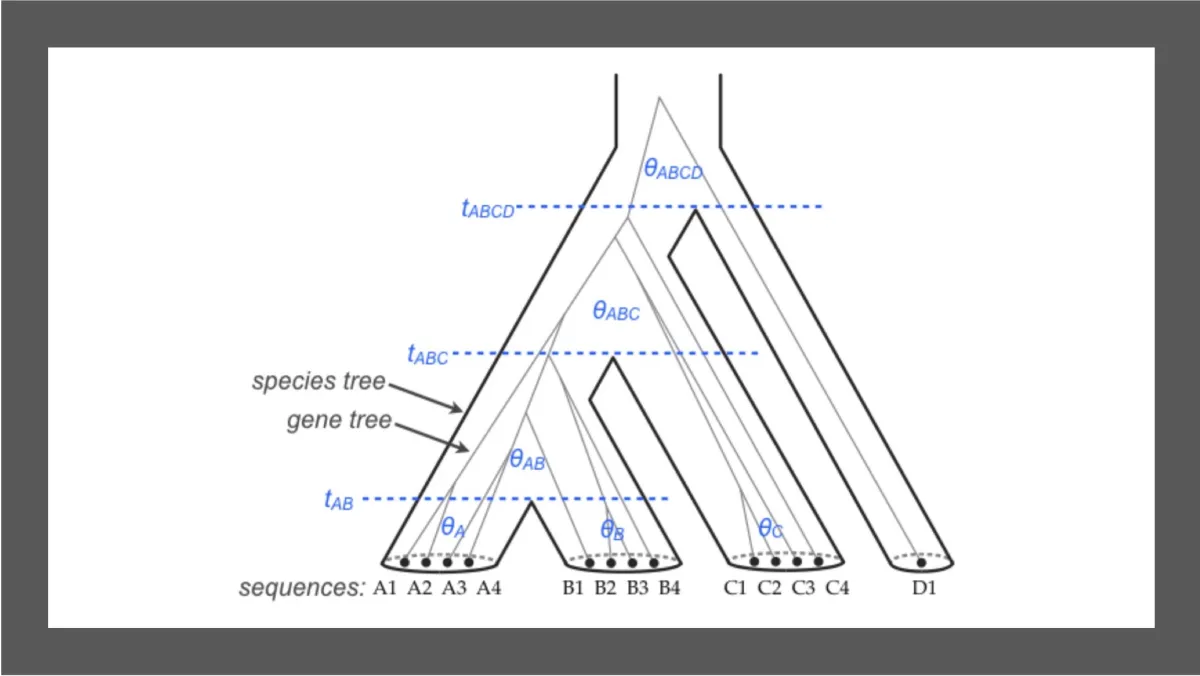
Workshop overview
Coalescent theory is a useful model for analyzing genetic data with applications in population genetics, phylogenetics, and species delimitation. During this full-day, in-person workshop you can expect a mixture of lectures, activities, and discussions. The activities will start simple and gradually increase in complexity. At each step, we will have time to discuss the results of your analyses as a group.
We will begin with an introduction to coalescent theory. Afterwards, you will have time for activities aimed at applying the model to real data. We will use a simple case of a single population and a single-locus dataset from humans to learn how the BPP program works and how to interpret results. Next, we will apply the model to multiple species with the multispecies coalescent (MSC) model using a bigger dataset containing multiple independent loci from the Great Apes. We will then add introgression and migration to the model. You will have a choice of datasets to explore (100s of genes from yeast or mosquitoes).
We will start the afternoon with a lecture and activity on species tree estimation using data from lizards. For this activity, there are two different datasets to analyze, and you can choose the one that most closely matches the characteristics of your own data. One of these is a phylogenomic dataset with an emphasis on species sampling, and the other is a phylogeographic dataset that emphasizes population sampling. The afternoon is dedicated to the controversial topic of species delimitation. We will begin with a group discussion on the issue before you use the MSC model to delimit species. For this exercise, you will work in groups to test species delimitation models using a simulated dataset. The correct answer will be revealed at the end of the exercise.
Coalescent theory
Activity 1: Coalescent in a single population - Human population data
Activity 2: Multispecies coalescent (MSC) - Great Ape data
MSC with introgression or migration
Activity: MSC-M (migration model) and MSC-I (introgression model) - Mosquitoes or yeast data
Species trees
Activity: Estimate species trees and associated parameters - Phylogeographic or phylogenomic data (lizards)
Species delimitation
Activity: Test species delimitation models - Simulated data
Software
Please bring your laptop with the software already installed. All operating systems are welcome. We will not have time on the day of the workshop to troubleshoot software installation problems. Full details will be emailed to registered participants the week before the workshop. Please get in touch if you haven't heard from us.
Registration
To assist us with workshop planning and catering please register by Wednesday 13 November.
Workshop feedback questionnaire here.
References
- Edwards, S. V., Z. Xi, A. Janke, B. C. Faircloth, J. E. McCormack, T. C. Glenn, B. Zhong, S. Wu, E. M. Lemmon, A. R. Lemmon, A. D. Leache, L. Liu, and C. C. Davis (2016). Implementing and testing the multispecies coalescent model: a valuable paradigm for phylogenomics. Molecular Phylogenetics and Evolution 94: 447–462.
- Flouri T, Jiao X, Rannala B, Yang Z. 2018. Species tree inference with BPP using genomic sequences and the multispecies coalescent. Mol Biol Evol 35:2585-2593.
- Flouri T, Rannala B, Yang Z. 2020. A tutorial on the use of BPP for species tree estimation and species delimitation. Pp. 5.6.1-16 in Scornavacca C, Delsuc F, and Galtier N, eds. Phylogenetics in the Genomic Era.
- Flouri T, Jiao X, Huang J, Rannala B, Yang Z. 2023. Efficient Bayesian inference under the multispecies coalescent with migration. PNAS120:e2310708120.
- Flouri T, Jiao X, Rannala B, Yang Z. 2020. A Bayesian implementation of the multispecies coalescent model with introgression for phylogenomic analysis. Mol Biol Evol 37:1211-1223.
- Hibbins MS, Hahn MW. 2022. Phylogenomic approaches to detecting and characterizing introgression. Genetics 220:10.1093/genetics/iyab1173.
- Huang J, Flouri T, Yang Z. (Huang2020 co-authors). 2020. A simulation study to examine the information content in phylogenomic datasets under the multispecies coalescent model. Mol. Biol. Evol. 37:3211-3224.
- Ji J, Jackson DJ, Leache AD, Yang Z. 2023. Power of Bayesian and heuristic tests to detect cross-species introgression with reference to gene flow in the Tamias quadrivittatus group of North American chipmunks. Syst Biol 72:446-465.
- Jiao X, Yang Z. 2021. Defining species when there is gene flow. Syst Biol 70:108–119.
- Leache, A. D. and M. K. Fujita (2010). Bayesian species delimitation in West African forest geckos (Hemidactylus fasciatus). Proceedings of the Royal Society of London B: Biological Sciences 277: 3071–3077.
- Leache, A. D. and B. Rannala (2011). The accuracy of species tree estimation under simulation: a comparison of methods. Systematic Biology 60: 126–137.
- Leache, A. D., R. B. Harris, B. Rannala, and Z. Yang (2014). The influence of gene flow on species tree estimation: a simulation study. Systematic Biology 63: 17–30.
- Leache, A. D., B. L. Banbury, J. Felsenstein, A. Nieto-Montes de Oca, and A. Stamatakis (2015). Short tree, long tree, right tree, wrong tree: new acquisition bias corrections for inferring SNP phylogenies. Systematic Biology 64: 1032–1047.
- Leache, A. D., T. Zhu, B. Rannala, and Z. Yang (2019). The spectre of too many species. Systematic Biology 68: 168–181.
- Leache, A. D. and J. R. Oaks (2017). The utility of single nucleotide polymorphism (SNP) data in phylogenetics. Annual Review of Ecology, Evolution, and Systematics 48: 69–84.
- Leache, A. D., H. R. Davis, S. Singhal, M. K. Fujita, M. E. Lahti, and K. R. Zamudio (2021). Phylogenomic assessment of biodiversity using a reference-based taxonomy: an example with Horned Lizards (Phrynosoma). Frontiers in Ecology and Evolution 9: 678110.
- Rannala, B., S. V. Edwards, A. D. Leache, and Z. Yang (2020). “The Multi-species Coalescent Model and Species Tree Inference”. In: Phylogenetics in the Genomic Era. Ed. by C. Scornavacca, F. Delsuc, and N. Galtier. No commercial publisher, Authors open access book, pp.3.3:1–3.3:21.
- Thawornwattana Y, Dalquen DA, Yang Z. 2018. Coalescent analysis of phylogenomic data confidently resolves the species relationships in the Anopheles gambiae species complex. Mol Biol Evol 35:2512-2527.
- Thawornwattana Y, Seixas FA, Mallet J, Yang Z. 2022. Full-likelihood genomic analysis clarifies a complex history of species divergence and introgression: the example of the erato-sara group of Heliconius butterflies. Syst Biol 71:1159-1177.
- Thawornwattana Y, Huang J, Flouris T, Mallet J, Yang Z. (Thawornwattana2023 co-authors). 2023. Inferring the direction of introgression using genomic sequence data. Mol. Biol. Evol. 40:msad178.
- Zhu T, Yang Z. 2021. Complexity of the simplest species tree problem. Mol Biol Evol. 10.1093/molbev/msab009
Leache Lab
Adam's visit is funded by the Centre for Biodiversity Analysis, hosted by Scott Keogh and Mitzy Pepper (ANU). Please get in touch if you would like to meet with Adam during his time in Canberra.
Location
Slatyer Seminar Room, 2nd floor, RN Robertson Building, Research School of Biology, Australian National University

