Trees and networks in evolutionary analysis: to tree or not to tree?
Systematists and macroevolutionary biologists naturally think of bifurcating trees as a natural way to investigate the evolutionary history of species, clades
Speakers
Event series
Content navigation
RegisterDescription
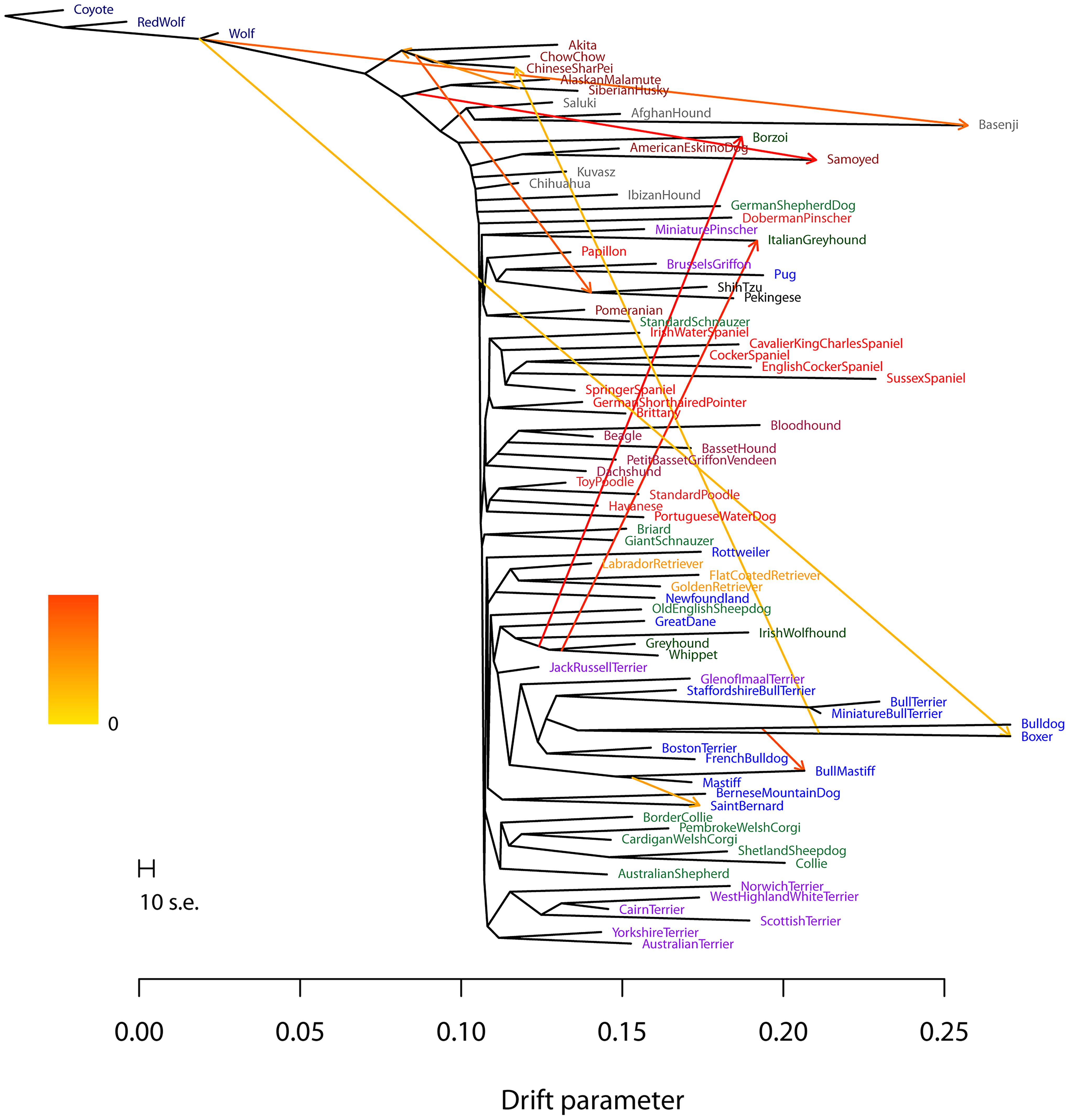
Systematists and macroevolutionary biologists naturally think of bifurcating trees as a natural way to investigate the evolutionary history of species, clades and assemblages.
However, from genomic comparisons there is growing evidence for reticulation - genetic introgression - among distantly, as well as closely related species and lineages.
Networks, long employed in ecological and other sciences, encompass bifurcating phylogenies - they are essentially phylogenies with connecting branches to allow for gene flow among taxa. Ignoring these reticulations results in potentially misleading estimation of phylogenies using species-tree methods, as well as misdiagnosis of species boundaries.
Although is now seems obvious that phylogenetics of related (e.g. congeneric) species should employ networks, the challenge is that the number of parameters to estimate rapidly exceeds data and/or computational limits as the number of OTUs increases.
A variety of heuristic and model-based approaches have been developed to model admixture across small numbers of taxa. These include simple ordination or genetic covariance methods (e.g. SpaceMix), through gene-tree or SNP based methods using summary statistics (Reich’s D, etc; TreeMix).
The most advanced methods in development are seeking to apply the “Multispecies Network Coalescent Model”, akin to the MSC model that underpins current species-tree methods (e.g. PhyloNet). Development and application of such methods, in place of traditional phylogenetic methods, will enrich our understanding of speciation and divergence histories of taxa.
Some background reading:
Edwards, et al. 2016. Reticulation, divergence, and the phylogeography– phylogenetics continuum PNAS 113(29).
Wen et al. 2016. Reticulate evolutionary history and extensive introgression in mosquito species revealed by phylogenetic network analysis PLoS Genetics 12(5).
People from all academic backgrounds and levels are welcome. Registration is free.
Hosted by the Centre for Biodiversity Analysis, TEA Talks (Techniques in Evolutionary Analysis) are a monthly series of short workshops that introduce a range of current methods and analytical approaches in phylogenetics, bioinformatics and macroevolution.
Location
Sciences Teaching Building Bldg 136, Linnaeus Way, Rm S2 (top floor), ANU
