Genome Diversity: Insights from long-read sequencing
The CBA and the National Centre for Indigenous Genomics are hosting a symposium on new advances in 3rd-generation genomics in genome assembly, haplotypes, pangenomes, and structural variation.
Speakers
Event series
Cost
Cost per person: 0.00
Content navigation
RegisterDescription
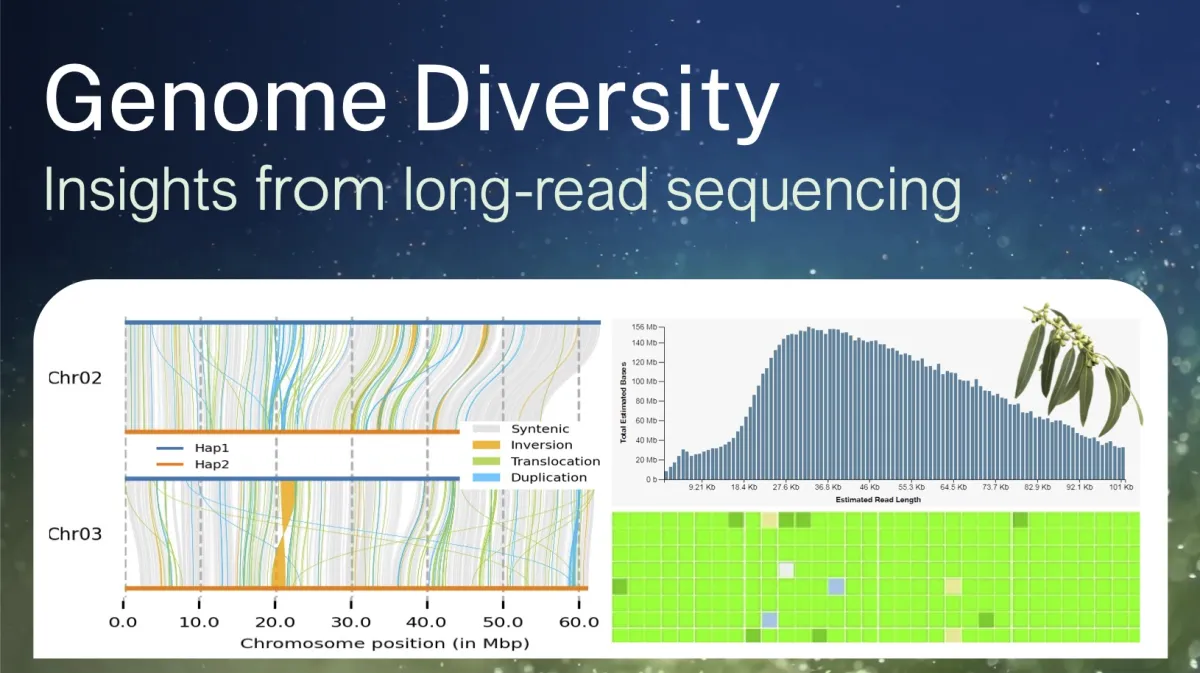
Building on Bioplatforms Australia initiatives (e.g. OMG, AusARG, TSI, NCIG) and parallel research, the Centre for Biodiversity Analysis and the National Centre for Indigenous Genomics will host a symposium on Genome Diversity: Insights from long read-sequencing. This will take place at ANU's Research School of Biology just ahead of the International Congress on Genetics and Genomics in Melbourne. We have taken advantage of several international visitors attending the Congress who are joining us in Canberra first. Together with leading local researchers, this symposium will be an exciting opportunity to hear from and network with others working in the field.
The meeting plans to showcase new advances in 3rd-generation genomics (PacBio and Nanopore) in genome assembly, haplotypes, pan-genomes, and structural variation. We will cover biological and technological insights and diversity among non-model populations of plants, animals, and humans. Multiple sessions will span topics across organisms and tools including genome repeats, methylation and other modifications that sit on top of the genome diversity. The program will link new insights from long read assemblies with biological function, phenotypic and adaptive diversity and highlight signatures of natural selection.
Download flyer
Registration and presentations
Thanks to the generous support provided by the CBA, NCIG, PacBio and Oxford Nanopore Technologies, we are pleased to offer free attendance to the symposium and social mixer. However, we ask you to please register for catering purposes, and let us know as soon as possible if you are unable to attend.
In the program we will have time for a small number of 10-min speed talks and space for poster presentations during the breaks and social mixer. We would particularly like to encourage students and ECRs to consider presenting as this is an excellent opportunity to share your ideas with leading international and local scientists.
On the registration form you can indicate if you would like to present a speed talk and/or poster, with space to include your title and a short abstract.
Registrations and abstract submissions close 30 June 2023.
Program
Download Program & Abstracts
Talks will be interspersed with plenty of time for discussion at tea breaks, lunches, a walk through the nearby Australian National Botanic Gardens, technological discussion panel, and an evening social mixer.
Thursday 13 July |
||
| 9:00 | Craig Moritz | Welcome |
| 9:15 | Talk: Justin Borevitz | Landscape (pan) genomics for climate adaptation in Eucalyptus |
| 9:45 | Talk: Sophie MacRae Orzechowski | Neo-sex chromosomes in Australian honeyeaters: a tale of gains and losses |
| 10:15 | Talk: Andy Bachler | Moth Pangenomes! And 3 long-read short-stories |
| 10:45 | Morning tea | Catcheside Court |
| 11:15 | Talk: Ann McCartney | The European Reference Genome Atlas: piloting a decentralized approach towards making biodiversity genomics equitable and inclusive |
| 11:45 | Talk: Scott Ferguson | Plant genome evolution in the genus Eucalyptus driven by structural rearrangements that promote sequence divergence |
| 12:15 | Speed talk: James Ferguson | Slow5: A fast and reliable format for nanopore sequencing |
| 12:25 | Speed talk: Floriaan Devloo Delva | High Throughput mitogenome sequencing to determine paternity of half sib pairs in a Close Kin Mark Recapture study |
| 12:35 | Speed talk: Benjamin Schwessinger | The short and the long. Nanopore sequencing beyond the genome. |
| 12:45 | Lunch | Catcheside Court |
| 1:15 | Break | Botanic Gardens walk |
| 2:45 | Talk: Karen Miga | Expanding studies of global genomic diversity with complete, telomere-to-telomere (T2T) assemblies |
| 3:15 | Talk: Qi Zhou | Evolution of chromosomes and their nuclear architectures of amniotes since their common ancestor |
| 3:45 | Talk: Rose Andrew | Disentangling the web of introgression in eucalypts |
| 4:15 | Break | 10-15 min |
| 4:30 | Technical panel discussion: David Hawkes, Tim Amos, James Ferguson, Ash Jones, Carolina Correa Ospina | Long-read sequencing platforms. Facilitated by Niccy Aitken. |
| 5:30 | Social mixer drinks & canapés | Catcheside Court |
| 6:30 | End of Day 1 | |
Friday 14 July |
||
| 9:00 | Talk: Scott Edwards | Pangenomes of North American Scrub-Jays (Aphelocoma) reveal abundant structural variation and rapid shifts in genome size |
| 9:30 | Talk: Jason Bragg | Genomics for plant conservation |
| 10:00 | Talk: Arthur Georges | Long-read sequencing technologies and sex determination -- on the cusp of a wave of new discovery? |
| 10:30 | Speed talk: Zixiong Zhuang | Eucalyptus viminalis structural variants |
| 10:40 | Speed talk: Agin Ravindran | Dissecting the impacts of gene fusion-mediated translational dysregulation in B-cell lymphoblastic leukemia to highlight novel therapeutic targets |
| 10:50 | Speed talk: Stephanie Chen | Conserving myrtle rust impacted species in the genomic era |
| 11:00 | Morning tea | Catcheside Court |
| 11:30 | Talk: Zander Myburg | Long-read sequencing of eucalypt hybrids: first insights into genome structural variation |
| 12:00 | Talk: Kate Farquharson | What can we achieve with a single long-read genome? Insights from two Extinct-in-the-Wild reptiles. |
| 12:30 | Talk: Hardip Patel | National Centre for Indigenous Genomics enabling the inclusion of First Nations peoples in genomics |
| 1:00 | Lunch | Catcheside Court |
| 2:00 | Symposium end |
Location
Robertson Lecture Theatre, Research School of Biology, RN Robertson Building, Australian National University, Canberra
Parking MAP
